Targeted protein Degraders
Target Protein Degraders (TPDs) represent an innovative class of small molecules designed to eliminate disease-causing proteins by harnessing the cell’s own degradation pathways, including the ubiquitin–proteasome and autophagy–lysosome systems. Over the past decade, new modalities have emerged — such as PROTACs (Proteolysis-Targeting Chimeras), molecular glues, CHAMPs (chaperone-mediated degraders), and LYTACs (lysosome-targeting chimeras) — that are reshaping drug discovery.
Unlike traditional inhibitors that merely block protein activity, TPDs provide a fast, reversible, and targeted chemical knock-down of disease-relevant proteins. This transformative approach extends druggable space beyond the 20% of the proteome accessible to conventional methods, opening the door to the remaining 80% of proteins once deemed “undruggable.”
At Axela Biosciences, our advanced TPD discovery platforms support a broad range of ligand classes and screening strategies. By combining high-content biology, robust assay systems, and translational expertise, we deliver cost-effective, reliable solutions to accelerate your TPD pipeline from early discovery through preclinical stages.
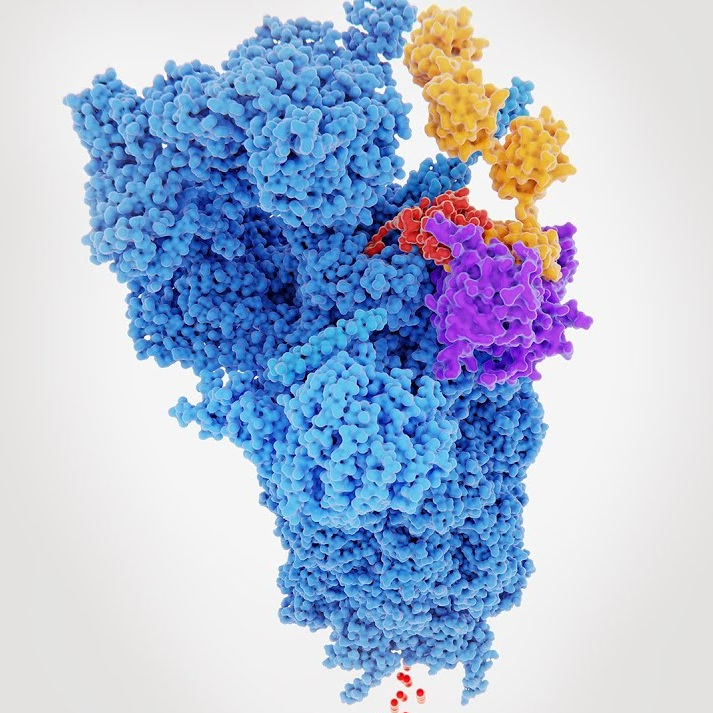
Structural model illustrating a targeted protein degrader (TPD) in action, bringing a disease-causing protein into proximity with the ubiquitin-proteasome system for controlled degradation.
Assay Platforms and Services
💠Quantitative protein degradation analysis (Simple Western, In-Cell Western, SDS-PAGE, Li-COR Imaging)
💠AlphaLISA and HTRF binding assays for CRBN, VHL, MDM2, BIR3, cIAP1, and more
💠NanoBRET assays for real-time E3 ligase engagement and ubiquitination kinetics
💠Ubiquitination detection with UbiQuant S ELISA and AlphaLISA
💠IHC and RPPA for translational biomarker analysis in tissues
💠High-throughput flow cytometry for protein degradation studies
💠Cell-based assays for permeability, thermal stability (CETSA), and viability
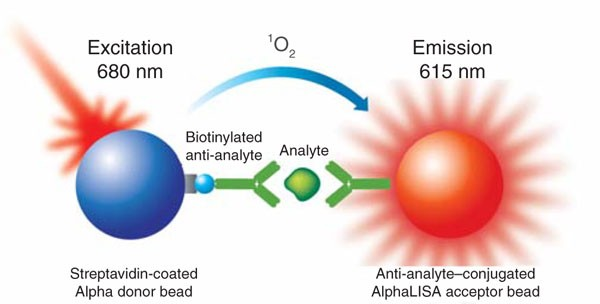
This image shows how the AlphaLISA assay works. When the target analyte brings the donor and acceptor beads together, light excitation at 680 nm triggers a bright signal at 615 nm. This simple, no-wash system enables highly sensitive detection of protein interactions, making it a powerful tool for drug discovery and biomarker research.
💠Identification and validation of novel ligands that reprogram E3 ligase specificity
💠PROTAC warhead development and DC50/Dmax quantification
💠Mechanistic insights into degrader efficacy using translational readouts (IHC, RPPA, and multiplex platforms)
💠Off-the-shelf assays including CRBN, BET-VHL binding, VHL and CRBN ternary complex formation
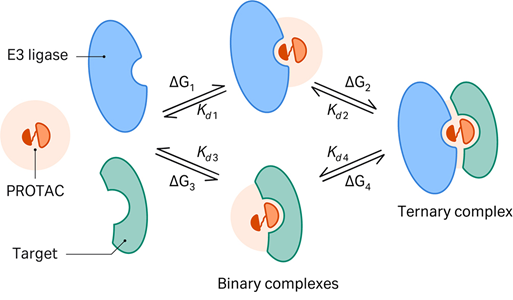
This illustration shows the mechanism of PROTAC-mediated protein degradation, where a PROTAC molecule brings together a target protein and an E3 ligase to form a ternary complex. This interaction tags the target protein for ubiquitination and subsequent degradation, offering a powerful strategy to eliminate disease-causing proteins that traditional drugs cannot address.
Example Data (a)
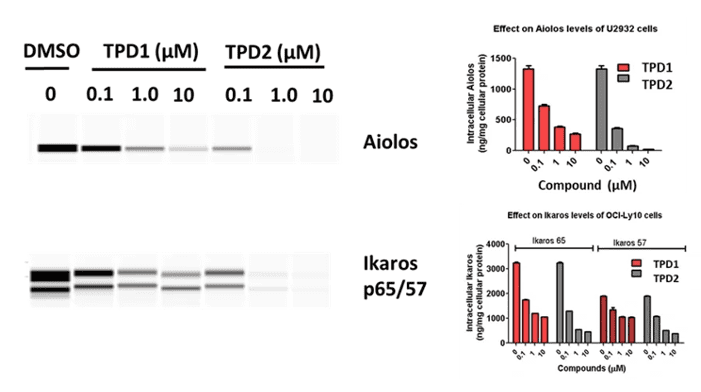
Example a: Simple Western
analysis demonstrating IKZF1/3 (Ikaros and Aiolos) degradation in human lymphoma cells treated with two different molecular glue compounds (TPD1 and TPD2). Dose-dependent reduction of target protein levels was observed, confirming compound efficacy in promoting protein degradation.
Example Data (b)
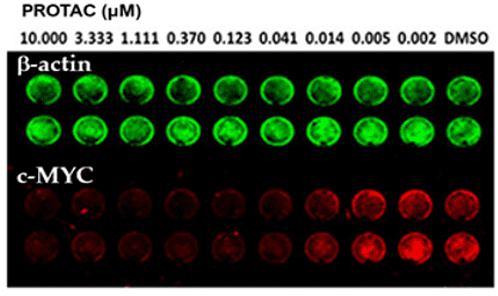
Example b: In-Cell Western
Quantification of c-MYC degradation in MV4-11 leukemia cells following treatment with a PROTAC compound. The assay demonstrates dose-dependent reduction of c-MYC protein levels, highlighting the compound’s ability to effectively induce target protein degradation inside cells.
Example Data (c)
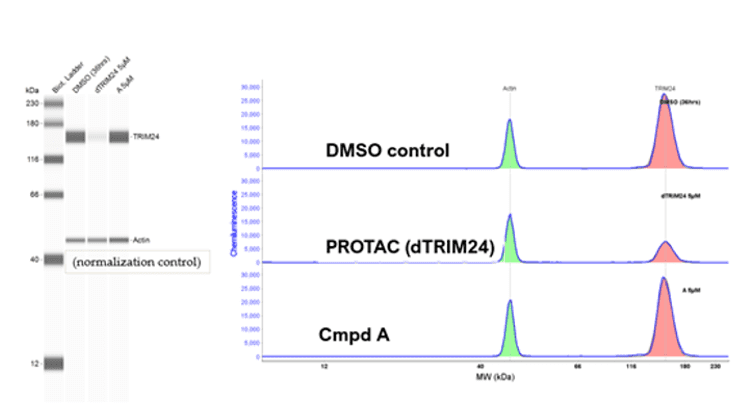
Example c: Simple Western
Assessment of TRIM24 degradation in MCF-7 human breast cancer cells treated with a PROTAC compound. The results demonstrate target-specific protein degradation, confirming the compound’s potential utility in breast cancer therapeutic development.
Example Data (d)
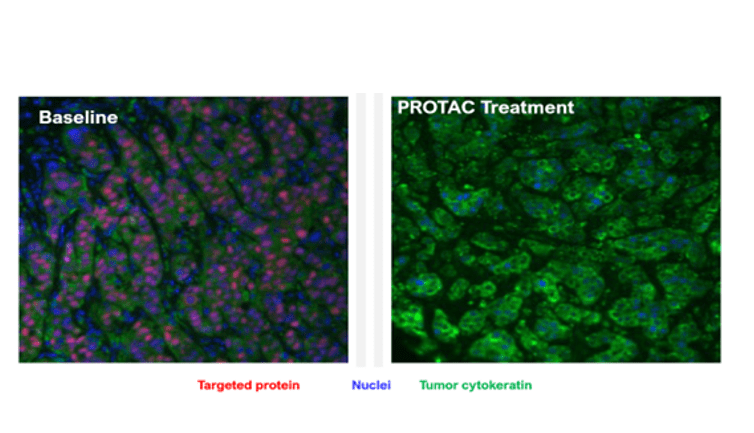
Example d: IHC Staining
Immunohistochemistry (IHC) showing protein degradation in tumor tissues from a xenograft model following PROTAC treatment, compared with baseline. Results highlight target-specific degradation within the tumor microenvironment.
Example Data (e)
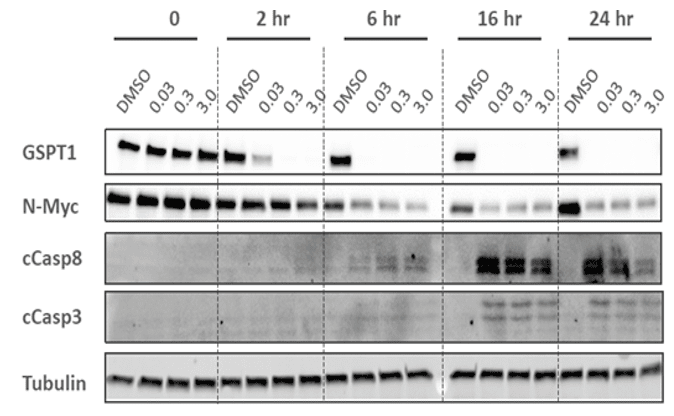
Example e: Traditional Western Blot
Analysis of GSPT1 degradation and caspase activation in cancer cells treated with a molecular glue compound. The time-course (2–24 hours) demonstrates protein degradation kinetics and downstream apoptotic signaling.
Example Data (f)
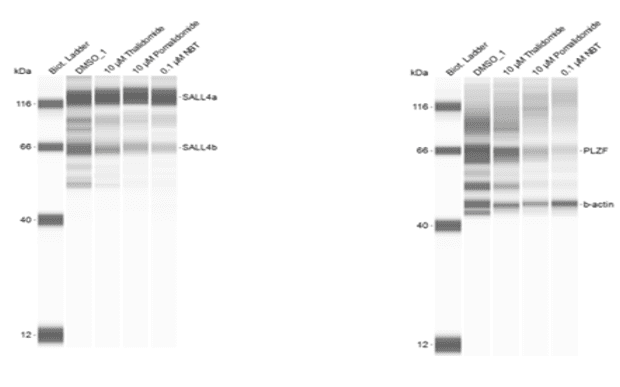
Example f: Simple Western
Quantification of SALL4 and PLZF degradation in induced pluripotent stem cells (iPSCs) treated with a CRBN-binding molecular glue compound. Both proteins are thalidomide-dependent cereblon neo-substrates and may play a role in drug-induced teratogenicity.